Setup
- Machine: 16-thread Xeon 2.6 GHz, 32 GB RAM, NVME PCIx16
- System: Ubuntu 16.04, Spark 2.4.4, Python 3.7.4, Pandas 0.25.1, Datatable 0.10.1
- Data: 100 million rows generated CSV (1.6 GB gzip compressed)
- Operation:
- Create a dataframe from a compressed file
- Group the dataframe by 3 columns
- Aggregate 2 different columns with 2 different function (group by: id1,id2,id3, aggregate: mean(v3), max(v1))
- Sort the result in descending order
- Get 10 first rows
Pandas
Script
python -c 'import time; start=time.time(); import pandas as pd; df2 = pd.read_csv("/home/agungw132/G1_1e8_1e2_0_0.csv.gz"); print(df2.groupby(["id1", "id2", "id3"]).agg({"v3": "mean", "v1": "max"}).sort_values(by=["v3", "v1"], ascending=False).head()); print(time.time()-start)'
Findings
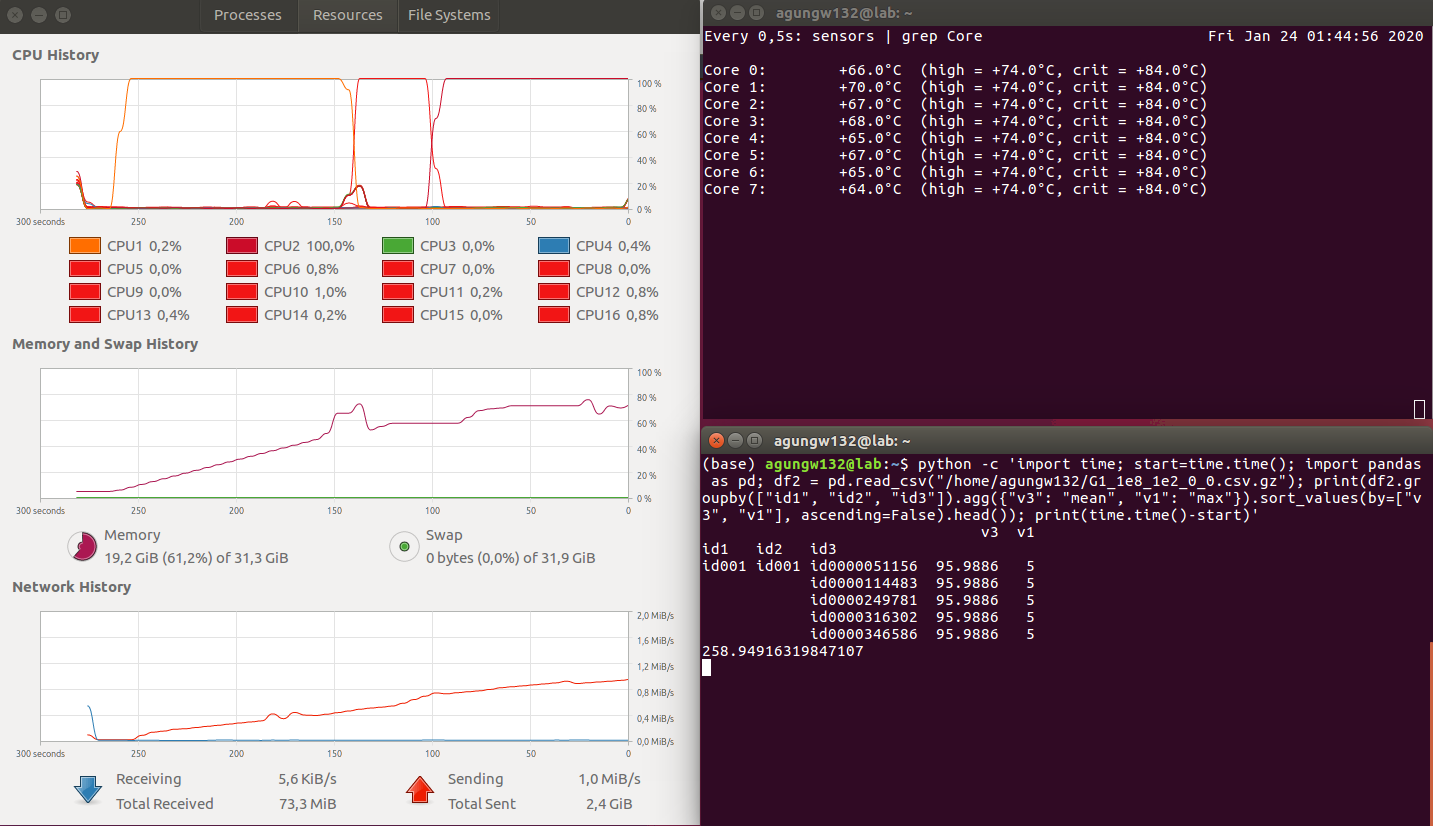
Datatable
Script
python -c 'import time; start=time.time(); import datatable as dt; d2 = dt.fread("/home/agungw132/G1_1e8_1e2_0_0.csv.gz"); print(d2[:,(dt.mean(dt.f.v3), dt.max(dt.f.v1)),dt.by(dt.f.id1, dt.f.id2, dt.f.id3)].sort(-dt.f.v3, -dt.f.v1).head()); print(time.time()-start)'
Findings
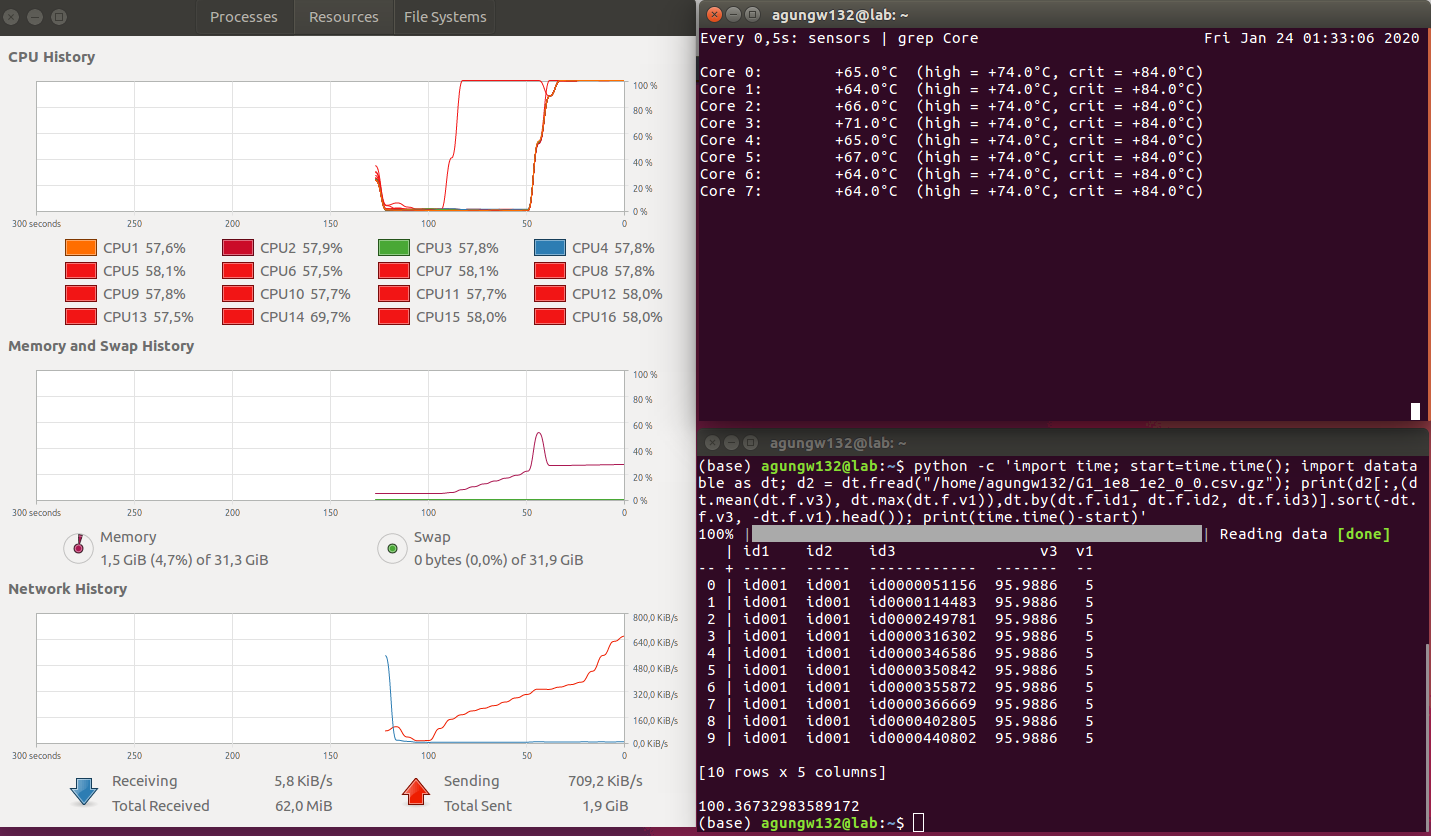
PySpark
Script
echo 'import time; start=time.time(); from pyspark.sql import SparkSession; spark = SparkSession.builder.appName("
trial").getOrCreate(); spark.sparkContext.
setLogLevel("ERROR"); d = spark.read.csv("/home/
agungw132/G1_1e8_1e2_0_0.csv.
gz", header=True); d.createOrReplaceTempView("d")
; print(spark.sql("SELECT * FROM (SELECT id1,id2,id3,avg(v3) as v3,sum(v1) as v1 FROM d GROUP BY id1,id2,id3) ORDER BY v3,v1 DESC LIMIT 10").collect()); print(time.time()-start)' | pyspark --master local[*] --executor-memory 32G --driver-memory 32G
Finding
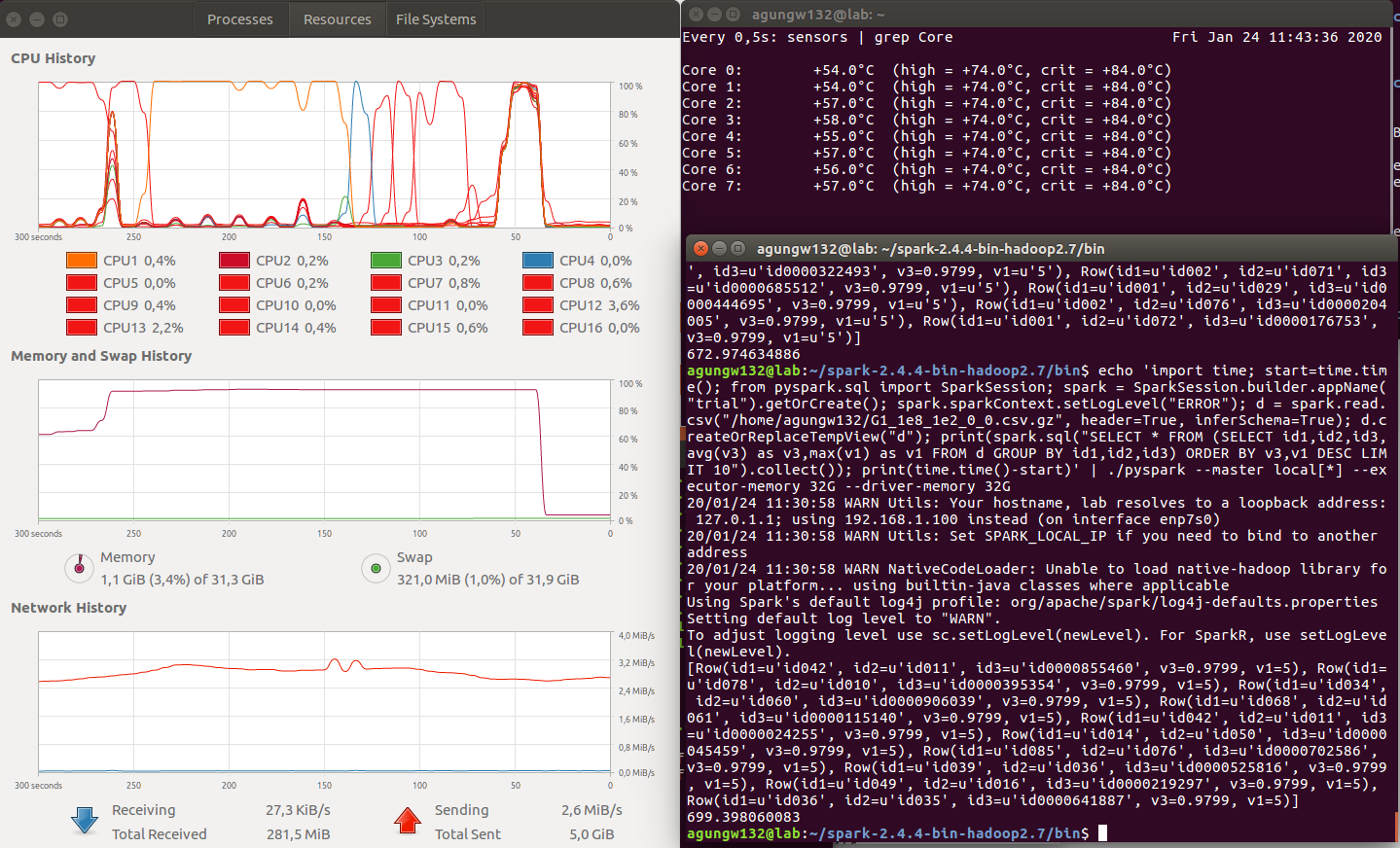
Discussion
- Execution Time (in seconds): Datatable (100) < Pandas (259) < Pyspark (699) (less is better)
- CPU Parallelism: Datatable > Pyspark > Pandas (bigger is better)
- Memory utilization: Pyspark > Pandas > Datatable (less is better)
The winner is obvious, i.e., datatable.